Warfarin Design
(→Class description) |
|||
| (54 intermediate revisions by 2 users not shown) | |||
| Line 45: | Line 45: | ||
[[image:WarfarinArc.JPG|frame|left|]] | [[image:WarfarinArc.JPG|frame|left|]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 92: | Line 111: | ||
== Uml diagram == | == Uml diagram == | ||
| + | |||
| + | |||
| Line 97: | Line 118: | ||
[[image:Umlnew.jpg|frame|left|]] | [[image:Umlnew.jpg|frame|left|]] | ||
| + | ==Initial design classes== | ||
| + | Domain logic is made out of 4 classes: | ||
| + | * ModelBuilder wraps up the logic for new model creation and the updates of existing models. It makes appropriate calls to WekaUtil and Datapoint Generators and data source as needed. Once the model is created and updated it invokes the data layer methods for persisting the model to the database. | ||
| + | * PresentationBuilder gives back the INR prediction. It uses DataPoint Generator to make the datapoint , and then it passes it to the WekaUtil to get the prediction for it. It also interprets Weka’s result depending on whether the predicted INR reading needs to be nominal or numeric. It is also responsible for making appropriate calls to data layer to persist (log) completed predictions. | ||
| + | * DataPointGenerator has the datapoint creation logic. It loops through the patient’s dose history and based on the attributes selected it calculates the amounts, comma separates them and puts them in to the datapoint. Its final product is an arff file (made of numerous datapoints) needed for model creation and updates as well as for making a prediction. | ||
| + | * WekaUtil process the API calls to Weka in order to physically create and update a model, persist it to the file, calculate the accuracy level of model created as well make a prediction on the generated datapoint. | ||
| + | Data Layer is primarily responsible for storing persistent data.It is made of one class - DataAccessHome. It communicates with an SQL server | ||
| + | database (reading and write data to) and xml file (reading data).The pattern used as a starting | ||
| + | point for this layer was Table Data Gateway pattern, where an object acts as a Gateway to a | ||
| + | database table. As this pattern suggests there should be an object for each physical table in | ||
| + | database where table data Gateaway “holds all the SQL for accessing a single table or view: | ||
| + | select, inserts, updates and deletes. Other code calls its methods for all interaction with the | ||
| + | database.” (Fowler 2003) | ||
| + | This pattern was slightly adapted when designing this prototype due to fact that our problem hasn’t | ||
| + | been addressed as big enough to have '''Table Data Gateway''' for each table in database, it was | ||
| + | considered that one Table Data Gateway(DataAccessHome) would be enough to handle all | ||
| + | methods for all table (as suggested for simple apps). | ||
| + | Due to the nature of the problem where we would be able to group all users’ actions into three | ||
| + | main streams: create model, update model or making a prediction. Those actions could then be | ||
| + | broken down into the many subroutines, where many of those would be shared between those | ||
| + | main action points. It then makes a perfect sense to organise our domain logic as suggested in | ||
| + | Transaction Script pattern. | ||
| + | A '''Transaction Script''' is essentially a procedure that takes input from the presentation, processes it | ||
| + | with validations and calculations, stores data in the database, and invokes any operations from | ||
| + | other systems.(Fowler,2003) | ||
| + | So it made a pefrect sence for me to use this pattern at the time. | ||
| + | ==The way the system currently works== | ||
| + | The way the system currently works is as follows: | ||
| + | # User(doctor) logs in. | ||
| + | # From here doctor has two options: It can either | ||
| + | #* Create a new model from a patient | ||
| + | #* Make a prediction for a patient | ||
| + | #;If he chooses to create a new model he needs to: | ||
| + | #* Select a patient | ||
| + | #* Select a type of the model he wants to create (single or multi patient model). If it is a single patient model only the chosen patient dose history would be used to “learn” a model. If the multi patient type is chosen the dose history for all patients would be used. | ||
| + | #* Next thing to do would be to select the model attributes that would be used to “learn” the model.When creating an attribute there are a number of different functions that can be applied to any numeric data field , such as sum,average and delta.Each function can be applied to different length time periods.Calculated attributes can be repeated to up to four times per example(for the current and previous three dosage intervals). General attributes (e.g. gender) can also be added or removed from the final group of attributes. Finally, there are potentially many different ML algorithms that can be used to create a model; the prototype uses WEKA code base as an source of ML algorithms and doctor can select which ones they wish to use for a given patient.The examples of some of the possible attributes are showing in the arff file below (eg. sumdose7day- meaning dose sum for the last 7 days starting from the last bloodtest taken) | ||
| + | #* Once all the above selection are made the user clicks on “CreateModel” button. This would invoke the CreateModel method of the ModelBuilder class passing the list of ModelAtttributes objects created by selecting the attributes from previous screen. The list of chosen algorithms would be passed as well as the patient the model is created for.CreateModel function would then create DataPointGenerator object and invoke appropriate methods to create the arff file based on patients doseage history and model attributes selected on the previous screen.Once the Arff file gets created CreateModel method would pass it’s location path as well as the selected algorithm names to WekaUtil object. WekaUtil makes appropriate API calls to WEKA’s machine learning algorithms and the model file gets saved on the disk(WekaUtil class), and persisted in the database(ModelBuilder class). | ||
| + | Arff file example: | ||
| + | [[image:arff.jpg|frame|left|]] | ||
| Line 164: | Line 224: | ||
| + | The sequence diagram for creating a model: | ||
| + | [[image:Sd1.jpg|frame|left|]] | ||
| Line 205: | Line 267: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 260: | Line 283: | ||
| + | #;If the user chooses to make a prediction: | ||
| + | #* Selects a patient to do prediction for | ||
| + | #* Selects a model to use for a prediction (can multi select) | ||
| + | #* A doctor needs to enter a new blood test results and also the new week long dose pattern that would like to get INR prediction for. If the doctor doesn't enter the desired pattern every 0.5 mg in the range of 0-15mg is used to predict on.Similare to creating a model (createModel method) we have GetPrediction method that invokes all needed actions in sequance.Starts with the GenerateDataPoint methods to get the Arff file for given dose pattern .Similare to creating a model, the arff file location gets passed to WekaUtil MakePrediction method. Lastly prediction gets persisted by DataAccessHome object. | ||
| + | = Design critique = | ||
| + | This is not an OO design. It looks more like procedural design. | ||
| + | Almost every possible heuristic is broken here, but let's just name few: | ||
| + | * [[Open/close principle]]. One of the main problems here for me personally is the lack of flexibility. When creating an arff file (formatted historical data used to "learn" a model) only limited set of functions(hardcoded) could be used to do so(sum, avg, delta, min, max).In the case when we would like to add a ratio as an a function to be used many parts of the code would need to be touched. The computation at the moment is done in DataPointGenerator by using ComputeValue(used for computing data using aggregate functions) and ComputeDeltaValue(used to compute delta value). To add a new way of computing would mean to add a new function that would perform a new computation(eg. ComputeRatioValue.This would also require a change of calling procedure to include a newly created function as one of the option of computing. This is clearly not desirable. | ||
| + | * [[Model the real world]] The existing design doesn't model the real world. Non of the classes are descriptive enough for a someone to understand what the classes responsibilities are. | ||
| + | * [[Keep related data and behavior in one place]] Data and behavior are not kept together. Model,ModelAttributes, Patient are only lazy classes ,carrying the data and don't have any behavior. | ||
| + | * [[Separation of concerns]] the program is not divided into small, logical, self explanatory classes. The classes mainly do too much .Example of the class doing too much is DataPointGenerator, which basically calculates all data needed writes them to the Arff file but also saves the file to the disk. | ||
| + | * [[Avoid god classes]] The DataPointGenerator is definitely a god class here. It has about 50% of functionality of whole application embedded in it. | ||
| + | * [[Long method smell]] The DataPointGenerator has the methods that involve many conditional statements, special in the part when calculating attributes values, this suggests that the method should be split up in the smaller tasks (methods).. | ||
| + | * [[Distribute system intelligence]] The breach of this principle is closely related to the [[Avoid god classes]] and [[Large class smell]]. I clearly didn't divide my system intelligence well enough. All system intelligence is concentrated in one class (DataPointGenerator). | ||
| + | * [[Coupling and cohesion]] There is quite a strong coupling between classes (eg. DataPointGenerator and ModelBuilder where ModelBuilder contains a pointer directly to a concrete class DataPointGenerator which provides the required behaviour). | ||
| + | = Improved design = | ||
| + | Following the smells and basic design principles I have come up with my improved design. The Uml follows: | ||
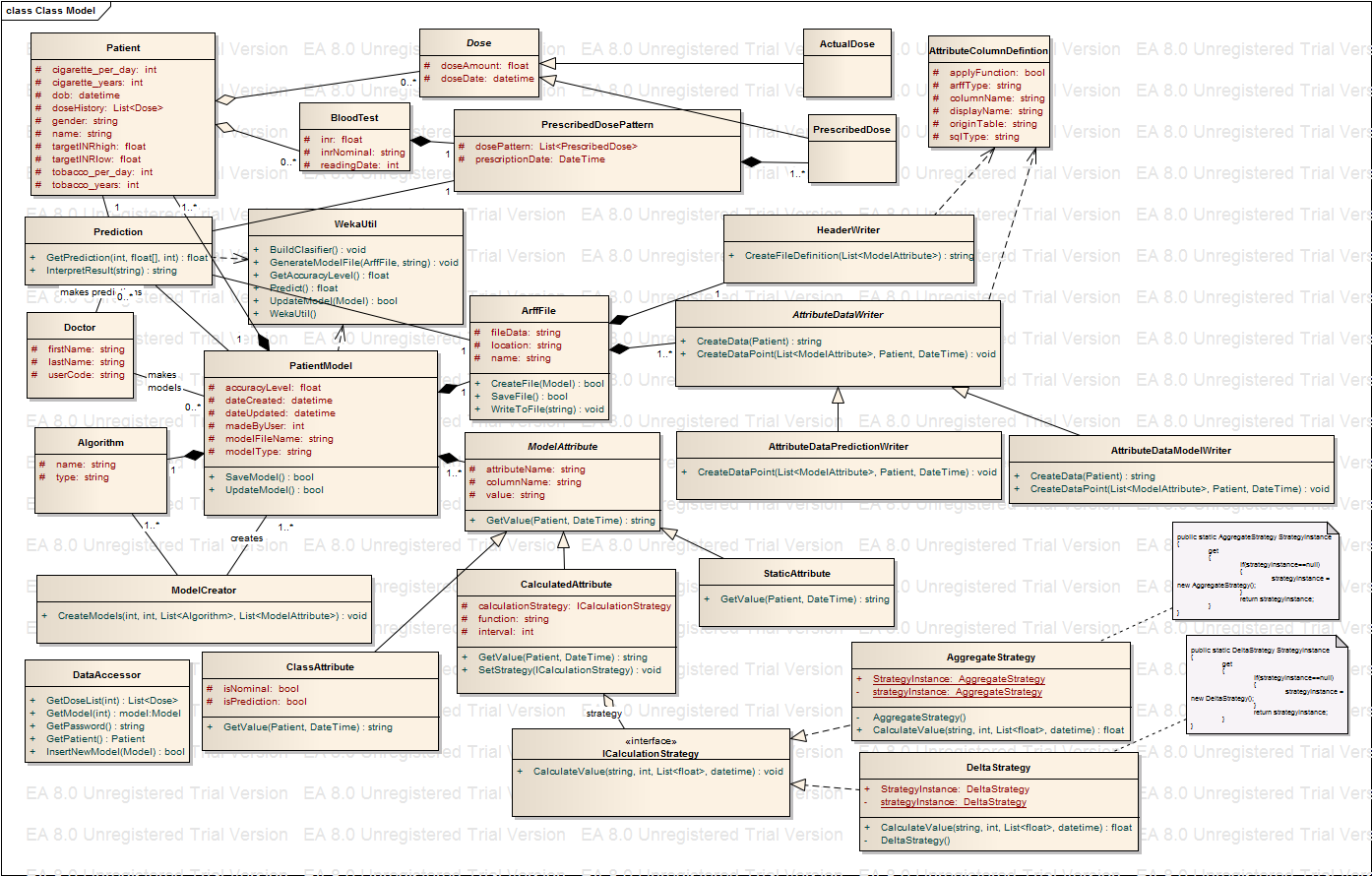
| + | [[image:WarfarinNew2.png|frame|left|]] | ||
| Line 304: | Line 345: | ||
| − | |||
| − | |||
| Line 354: | Line 393: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | = Summary = | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | = New design analyze = | ||
| + | Getting to the stage when I am comfortable with my new design involved many hours of grappling with some important design issues and decisions. In that process I have been through close to 10 different UML designs. Although I tried to follow the design best practices and maxims, sometimes compromises had to be made in terms of the design approach in the given situation. Not an easy task. | ||
| + | I will first name all my classes and briefly explain what their roles are. Then I will talk about principles that I strongly followed, conflicts that I encountered, and how and why I made the decisions that I made. | ||
| + | Before I list all my classes I want to point out that my UML has most important fields, but because I tried to keep UML complete but not cluttered I left out constructors and public properties (getters and setters). I also removed all fields that are already implied by relationships. | ||
| + | |||
| + | ==Classes used in the new design== | ||
| + | |||
| + | * '''Patient''' - this class is used to present the patient with it's history. It is mostly data class, used to carry data relevant to a patient. Saving the patient, updating the patient or finding the patient (or list of patients) are all implemented in DataAcessor class, due to layered architecture that I wanted to achieve here. Will talk about this more in the next section. | ||
| + | * '''BloodTest''' - this class is used to carry data related to regular blood tests that person takes. Every person could have many of those. | ||
| + | * '''Dose''' - this abstract class presents one dose on the specific date. People take Warfarin once a day , the time was not relevant as per initial requirements but this could change so the data type used here is DateTime (not just date). Each dose has two concrete subclasses ActualDose and PrescribedDose. Ideally those should be the same but there is a good chance to be different due to human error. | ||
| + | * '''Doctor''' - this class presents a doctor that is our design same as a user that logs to the website. This is the reason for having a user code field. The doctor creates a PatientModel and Prediction. | ||
| + | * '''PatientModel''' - this class represents a model that can be used to predict INR reading. I is responsible for constructing a model, saving it and updating it. | ||
| + | * '''Prediction''' - this class is responsible of making a prediction based on the model selected by a user. It interacts with Arff file and model and based on the dose pattern that doctor find appropriate makes a prediction. It also makes a calls to WekaUtil in order to utilize matching learning algorithms. | ||
| + | * '''Algorithm''' - this class is again data class used to present one ML algorithm. Every model is build by using one. | ||
| + | * '''ModelCreator''' - this class has responsablity of iterating through selected list of algorithms for a selected list of attributes and create as many models as there are algorithms. | ||
| + | * '''ArffFile''' - this class represents the Arff file containing patients history data transformed into data points as per selected model attributes. It is responsible for creating the file with all it's data and saving it to the disk. | ||
| + | * '''HeaderWriter''' - this class is used to create arff file header definition. | ||
| + | * '''AttributeDataWriter''' - this is an abstarct class subclassed into '''AttributePredictionDataWriter''' and '''AttributeModelDataWriter'''. Both make data points in arff file where the first one does it for making a prediction while the subsequent one makes it for model creation. | ||
| + | * '''ModelAttribute''' - this is an abstract class responsible of representing a model attribute created by a user.Important fields here are attributename but also the columnname which reporesents the name of the data column(database column) whose data this attribute should be based on. This column should match one of the patient's class fields .Value represents the calculated model attribute value for a given patient and given bloodtest date. | ||
| + | * '''ClassAtribute''' - this is ModelAttribute's subclass. It can be nominal or numeric but also differs if is made for prediction or model building purposes. | ||
| + | * '''CalculatedAttribute''' - this a MOdelAttribute sublass that usesfunction passed to it to invoke a sutable strategy to claculate the attribute value. | ||
| + | * '''StaticAttribute''' - this is the concrete class, ModelAttribute subclass used to give back the information on the patient that doesn't need calculation (eg.dob,gender...) | ||
| + | * '''ICalculationStrategy''' - this is an interface used to specify the strategy signiture.. Is then implemented by two concrete strategies. | ||
| + | * '''AggregateStartegy''' - this class reporesents a concrete strategy taht implements ICalculationStrategy used to calculate modelAttribute value when there is an aggregate function to be used (sum,min, max,avg). | ||
| + | * '''DeltaStrategy''' - a concrete class that implements ICalculationAttribute and calculates delta value between sum of column data for two subsequent blood test readings. | ||
| + | * '''AttributeColumnDefinition''' - this class hold all necessary information on patient's database column. It is a datacolumn that is open for user to select when designing a model attribute. Therefore we need to know it's database characteristics but also which data field in Arff file this data field match to.It is used by '''HeaderWriter''' when creating the arff file header but also by '''AttributeDataWriter''' | ||
| + | * '''WekaUtil''' - this class is used to make all API calls to WEKA algorithms. | ||
| + | * '''DataAccessor''' - this class calls SQL stored procedures for retrieving, updating and persisting the data classes. | ||
| + | |||
| + | |||
| + | ==Design decisions== | ||
| + | * ActualDose and PrescribedDose classes are currently empty. There is nothing to distinguish them at this point so it might look odd to have them .Few principles work against them [[You ain't gonna need it]] , [[Eliminate irrelevant classes]] and [[Lazy class smell]]. I decided to include them because [[Model the real world]] principle would recommend to have them as the difference between the two are bound to happen in real life but also because I will need to do the investigation on how much actual dose can vary to prescribed dose without causing an undesirable INR reading. | ||
| + | |||
| + | * PatientModel object can be made based on a single patient history and in that case would only make sense to be used by that patient, but if it was made on multi patient history can be used in prediction for any patient. So the principle [[Model the real world]] would suggest here for me to have two subclasses here '''SinglePatientModel''' and '''MultiPatientModel''' and make my model more extensible , but I think that [[You ain't gonna need it]] probably overrules this in this case. I think that having a list of patient field on the PatientModel that can have either one element in a case of single model or many patients in a case of a multi model would be sufficient for now. If I ever need to find if a patient's history was included in the model I can add the method IsIncluded() that would do that kind of testing for me. | ||
| + | |||
| + | * I had a problem with finding the right names for a few classes. One is PatientModel class. I initially named it Model but decided that the name is too generic. I was thinking about '''Classifier''' as another option, but only the person that is familiar with the domain can understand it otherwise the term could be too vague. So I made a decision to call it '''PatientModel''' since it is made on the bases of patient data and it is made for the purpose of predicting the patient’s blood test, so it is all to do with the patient. The other one is the '''ModelAttribute''' class. It is also pretty vague and not descriptive but is used in domain and I think I have used the term quite a lot in my discussion of how the system works that even a new comer can find it comprehensive. | ||
| + | |||
| + | * Initially in my old design I had a class named '''ModelBuilder''' that iterated through the chosen algorithms and made many new models based on one arff file ( so based on the same patient history) .I initially thought that PatientModel itself might do this by changing the algorithm field when iterating through the list of algorithms selected and since all the other characteristics of the PatientModel would stay the same I would just call WekaUtil from PatientModel to create as many model files as there are algorithms. But PatientModel shouldn't really take care of this. This would break [[Single Responsibility Principle]] and [[One key abstraction]]. So I introduced ModelCreator and pulled only CreateModel method out into this new class. This method should create a completely new model each time (call the PatientModel constructor) and set the correct parameters. The PatientModel would then have a responsibility of representing a model and not creating lots of new ones. | ||
| + | |||
| + | * I have some data classes in my design (e.g. patient and algorithm) so I think I should justify them. The [[Lazy class smell]] and [[Data class smell]], as well as the [[Anaemic domain model]] anti-pattern apply here. However, I think that [[Model the real world]] was important in this instance and it is highly likely that more behaviour will be added to the classes later. | ||
| + | |||
| + | * I already mentioned when I was criticising my initial design that it didn't follow [[Open-closed principle]] because it didn't allow introducing a new way of calculating the modelAttribute without making some serious changes to DataPointGenerator. In order to change this and make this design extensible I introduced '''Strategy pattern''' when it comes to calculating the attribute values. Since I have 3 different types of ModelAttributes and only CalculatedAttribute needs a computationStrategy, I made CalculatedAttribute a context for my strategies. Depending on the selected calculated attribute (the function used) an appropriate strategy would be instantiated. I also made strategies Singletons to make sure that I am not needlessly instantiating a new strategy objects for each datapoint and for the cases when few attributes are using same strategy (and the only difference for example is interval to apply aggregate function to). | ||
| + | |||
| + | * There is possibility for implementing one more design pattern in this design. The Template pattern. I thought that I could use it in the area of AttributeWriter where I need to make a different files depending on weather the file is made for prediction or for building a model. I could have an abstract class that would define the steps in writing header and data to the arfffile and have methods like | ||
| + | WriteHeading() | ||
| + | WriteAttributeValues() | ||
| + | and then implement those in subclasses differently. I decided not to do this since there are not as many different steps in writing data to the file (it is only one, header would be the same in both cases, and it is a common knowledge that if pattern doesn't help with code flexibility a great deal it is better not to implement it since it is adding a complexity. | ||
| + | |||
| + | * When it comes to encapsulation I investigated couple of relevant but conflicting design principles: [[Hide data within its class]] and [[Avoid protected data]].I also read [[Encapsulation boundary]] research and decided to use object encapsulation. I am aware that this is going to break Riel's heuristic of hiding data within its class, but it only seemed natural to go with object encapsulation and allow subclasses to access parent's fields by making them protected. Those fields would remain private for the rest of the world. Since my project is done in C# protected fields are available to use. | ||
| + | |||
| + | * I would like to explain my decision on putting all the database methods into one common class called DataAcessor. I am aware that this is breaking a very important OO principle [[Keep related data and behavior in one place]], but this would on the other hand go well with other conflicting principle [[Separation of concerns]] since writing to the database is not really the functionality of the Patient object or PatientModel object for example, so this is something that needs to be separated out. I decided to obey [[Separation of concerns]] here for one more reason. Having a database access all in one place makes the data source change doable by changing only one class. This would mean that I implemented n - tier architecture (having presentation layer, domain logic layer , data access layer and physical database layer). If my application would to grow much bigger then what it currently is, my class DataAccessor would become too big and I would end up with [[Large class smell]] but for now it seems like a good decision backed up with the reasoning that we don't expect this to grow very much at all. | ||
| + | |||
| + | == Design extensibility == | ||
| + | |||
| + | Easy extensibility and flexibility of the design were high on my priority list when I started improving my initial design. I found really crucial to be able add new types of model algorithms to the process of "learning" a model. Those could come form collecting a new data on the patients that we thought could make a difference to the prediction results ( eg. we could decide to record and process blood pressure) but also we could think of better ways of transforming patient's history (eg. like using the data ratios). | ||
| + | I think that the design that I had initially would require for every class to be changed to achieve that request. This is naturally not desirable thing to have. | ||
| + | In the new design I could achieve adding a new patient data by adding a new record to the XML file that is used to feed AttributeColumnDefinition class. This would bring this new field to the user selection screen. I would also need to add that same field to the patient class. | ||
| + | To add a new way of computing any numeric values stored on a patient , I would only need to add a new strategy with CalculateValue method implementation. By doing this I am not changing the existing classes, I am only extending the design by adding a new class that is inheriting from abstract class that didn't need to change. This makes [[Open-close principle]] and [[Stable abstractions principle]] fully implemented. | ||
| + | |||
| + | =Summary= | ||
| + | |||
| + | To summerize I am think taht this project was very useful. The work that I have done on redisagning my code made me rethink and question every single idea that I initially had, which overallhelped me grow in OO sense. I have started with a very messy code that has most of the required functionality but would be an nightmare to maintain and extend. I think that the quote that we shouldn't be programming just to achieve the functionality but for other programmers to continue the work on and extend, makes a lot of sense . If everyone would think in that way we would not just spend less time doing maintaince but would save the money spent on the projects overall. I expect that anyone working on my project from this point on should find it easy to work with and quite intuitive . | ||
| + | |||
| + | =Code= | ||
| + | |||
| + | This is my inital code [[Media:WarfarinPredictor.zip]] | ||
| + | <br> | ||
| + | Note: all the weka.dlls that are used to make API calls to weka's machine learning algorithms are taken out due to the size ( over 8MB) | ||
| + | <br> | ||
| + | Code for the improved design is coming soon. | ||
Latest revision as of 01:57, 25 November 2010
Contents |
Background
Warfarin predictor is a result of my cosc366 research project. It is a web based application that uses machine learning algorithms to predict a right dose for heart-valve transplant patients.
What is Warfarin?
Warfarin is taken by patients as a blood thinner in order to prevent blood clotting In It is very important that right dose of Warfarin has been taken so that patient has a minimal chance of clotting, while still ensuring that patient has enough clotting ability so that he or she does not bleed to death. Blood tests needs to be performed in order to know if INR (International Normalised Range, a normalised measure of the time taken for blood to clot) reading for the patient is within a safe range. The target INR range differs depending on the type of valve replacement, and to some degree and also on the patient. The way the patient will react to Warfarin intake depends on many factors. These include an individual's age, weight and gender, lifestyle habits such as alcohol and tobacco consumption, and even environmental factors. So how to help doctors prescribe the right dose of Warfarin? It is hoped that modern technology particularly a machine learning solution could be the way to go. So this is where my journey starts.
Project goal
The main goal of this research is to build on what is learned by in now in domain of using a machine learning to prescribe Warfarin, and develop a prototype for web-based Warfarin Predictor. This prototype will need to allow for a doctor to have enough flexibility to create and combine different attributes (information that a doctor would find valuable) in order to create a model that doctor can then use to predict Warfarin intake of the patient with the greatest accuracy. By giving doctor enough flexibility to self create the input attributes and combine them in order to make a prediction model. It is hoped that it would be possible to find the combination of attributes and algorithm that could best suit individual patient. This could be particularly important when the patient does not have a long history of taking Warfarin and other patient’s history is to be used. The web-based prototype is not going to give one best solution for all it would act more as a tool that is going to be used by doctors to help them make a right decision. It is hoped it may improve accuracy of dosage, cut costs, decrease physician workload and help with keeping an accurate patient medical history. The other goal of this research is to do some experimental work once the web-based predictor is created to give some insight to what type of attributes should work best with which machine learning algorithm as it is expected that some combination would not give any value while others could lift the accuracy of Warfarin prediction.
Requirements
Initial Requirements
- System users: administrators, doctors.
- Administrators can add new doctors.
- Doctors adds patients, patient records.
- Main use case is to record a new blood test (INR reading), get a dose prediction.
- Dose predictions may be returned in several ways, probably not advisable to just give the dose to prescribe because of liability issues. For example, could give a graphical representation of the likelihood of being below the safe range, in range or above.
- The system needs to be able to "learn" a model from the historical data. This involves transforming the data into a special format (an "ARFF" file) and submitting this to the learning algorithm.
- The data-to-arff step needs to be flexible (i.e. able to be easily changed if the learner's data requirements change.
- Models will need to be persisted.
- The data needs to be reliably persisted.
Future requirements
- There may be a need to record other data for the patient, possibly temporally, such as diet, other drugs taken, smoking and alcohol consumption.
- Generate graphs from multiple classifiers (predictions coming from different machine algoritm) to present to the doctor
- Investigate other ensemble methods (combine the prediction coming from different machine learning)
- Investigate using more classes (e.g. not just high and low but also very high and very low)
Current system design
Initial design
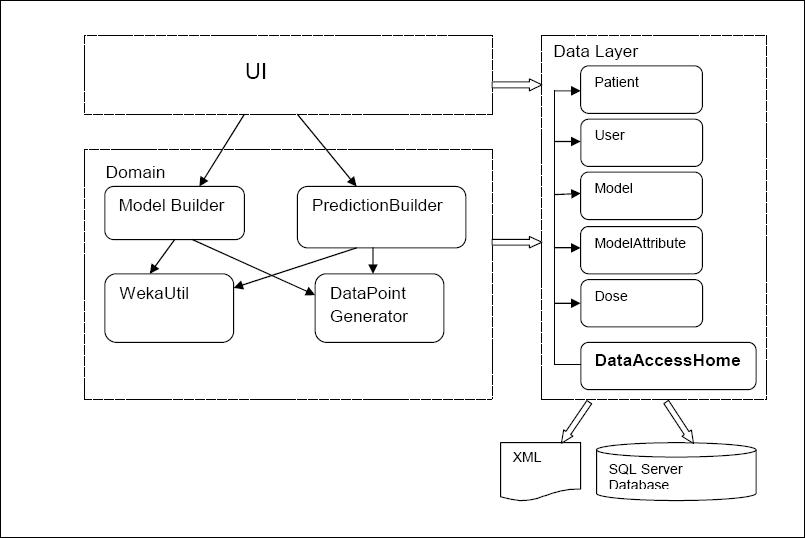
The high level presentation of classes used and their mutual interaction
Uml diagram
Initial design classes
Domain logic is made out of 4 classes:
- ModelBuilder wraps up the logic for new model creation and the updates of existing models. It makes appropriate calls to WekaUtil and Datapoint Generators and data source as needed. Once the model is created and updated it invokes the data layer methods for persisting the model to the database.
- PresentationBuilder gives back the INR prediction. It uses DataPoint Generator to make the datapoint , and then it passes it to the WekaUtil to get the prediction for it. It also interprets Weka’s result depending on whether the predicted INR reading needs to be nominal or numeric. It is also responsible for making appropriate calls to data layer to persist (log) completed predictions.
- DataPointGenerator has the datapoint creation logic. It loops through the patient’s dose history and based on the attributes selected it calculates the amounts, comma separates them and puts them in to the datapoint. Its final product is an arff file (made of numerous datapoints) needed for model creation and updates as well as for making a prediction.
- WekaUtil process the API calls to Weka in order to physically create and update a model, persist it to the file, calculate the accuracy level of model created as well make a prediction on the generated datapoint.
Data Layer is primarily responsible for storing persistent data.It is made of one class - DataAccessHome. It communicates with an SQL server database (reading and write data to) and xml file (reading data).The pattern used as a starting point for this layer was Table Data Gateway pattern, where an object acts as a Gateway to a database table. As this pattern suggests there should be an object for each physical table in database where table data Gateaway “holds all the SQL for accessing a single table or view: select, inserts, updates and deletes. Other code calls its methods for all interaction with the database.” (Fowler 2003) This pattern was slightly adapted when designing this prototype due to fact that our problem hasn’t been addressed as big enough to have Table Data Gateway for each table in database, it was considered that one Table Data Gateway(DataAccessHome) would be enough to handle all methods for all table (as suggested for simple apps).
Due to the nature of the problem where we would be able to group all users’ actions into three main streams: create model, update model or making a prediction. Those actions could then be broken down into the many subroutines, where many of those would be shared between those main action points. It then makes a perfect sense to organise our domain logic as suggested in Transaction Script pattern. A Transaction Script is essentially a procedure that takes input from the presentation, processes it with validations and calculations, stores data in the database, and invokes any operations from other systems.(Fowler,2003) So it made a pefrect sence for me to use this pattern at the time.
The way the system currently works
The way the system currently works is as follows:
- User(doctor) logs in.
- From here doctor has two options: It can either
- Create a new model from a patient
- Make a prediction for a patient
- If he chooses to create a new model he needs to
- Select a patient
- Select a type of the model he wants to create (single or multi patient model). If it is a single patient model only the chosen patient dose history would be used to “learn” a model. If the multi patient type is chosen the dose history for all patients would be used.
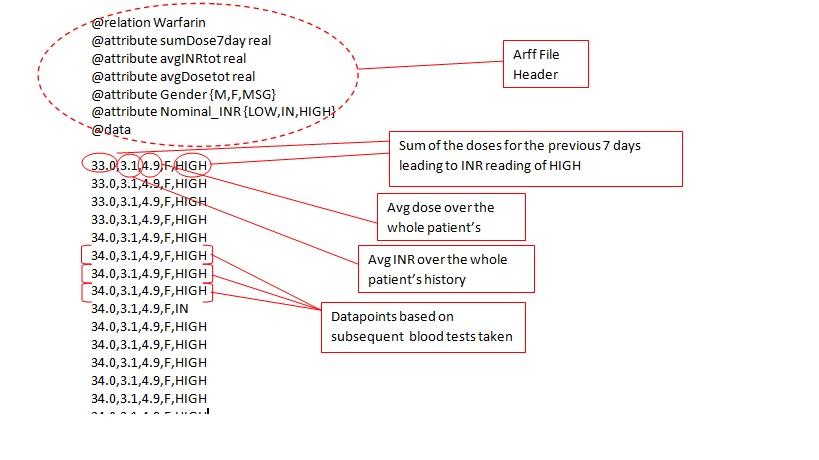
- Next thing to do would be to select the model attributes that would be used to “learn” the model.When creating an attribute there are a number of different functions that can be applied to any numeric data field , such as sum,average and delta.Each function can be applied to different length time periods.Calculated attributes can be repeated to up to four times per example(for the current and previous three dosage intervals). General attributes (e.g. gender) can also be added or removed from the final group of attributes. Finally, there are potentially many different ML algorithms that can be used to create a model; the prototype uses WEKA code base as an source of ML algorithms and doctor can select which ones they wish to use for a given patient.The examples of some of the possible attributes are showing in the arff file below (eg. sumdose7day- meaning dose sum for the last 7 days starting from the last bloodtest taken)
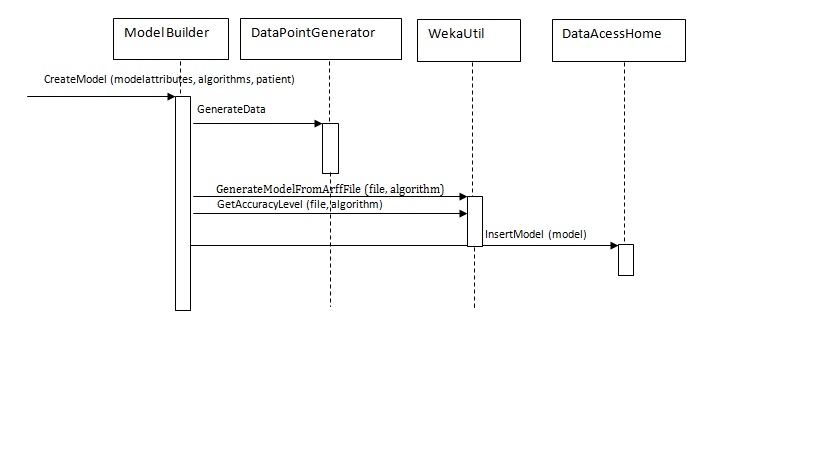
- Once all the above selection are made the user clicks on “CreateModel” button. This would invoke the CreateModel method of the ModelBuilder class passing the list of ModelAtttributes objects created by selecting the attributes from previous screen. The list of chosen algorithms would be passed as well as the patient the model is created for.CreateModel function would then create DataPointGenerator object and invoke appropriate methods to create the arff file based on patients doseage history and model attributes selected on the previous screen.Once the Arff file gets created CreateModel method would pass it’s location path as well as the selected algorithm names to WekaUtil object. WekaUtil makes appropriate API calls to WEKA’s machine learning algorithms and the model file gets saved on the disk(WekaUtil class), and persisted in the database(ModelBuilder class).
Arff file example:
The sequence diagram for creating a model:
- If the user chooses to make a prediction
- Selects a patient to do prediction for
- Selects a model to use for a prediction (can multi select)
- A doctor needs to enter a new blood test results and also the new week long dose pattern that would like to get INR prediction for. If the doctor doesn't enter the desired pattern every 0.5 mg in the range of 0-15mg is used to predict on.Similare to creating a model (createModel method) we have GetPrediction method that invokes all needed actions in sequance.Starts with the GenerateDataPoint methods to get the Arff file for given dose pattern .Similare to creating a model, the arff file location gets passed to WekaUtil MakePrediction method. Lastly prediction gets persisted by DataAccessHome object.
Design critique
This is not an OO design. It looks more like procedural design. Almost every possible heuristic is broken here, but let's just name few:
- Open/close principle. One of the main problems here for me personally is the lack of flexibility. When creating an arff file (formatted historical data used to "learn" a model) only limited set of functions(hardcoded) could be used to do so(sum, avg, delta, min, max).In the case when we would like to add a ratio as an a function to be used many parts of the code would need to be touched. The computation at the moment is done in DataPointGenerator by using ComputeValue(used for computing data using aggregate functions) and ComputeDeltaValue(used to compute delta value). To add a new way of computing would mean to add a new function that would perform a new computation(eg. ComputeRatioValue.This would also require a change of calling procedure to include a newly created function as one of the option of computing. This is clearly not desirable.
- Model the real world The existing design doesn't model the real world. Non of the classes are descriptive enough for a someone to understand what the classes responsibilities are.
- Keep related data and behavior in one place Data and behavior are not kept together. Model,ModelAttributes, Patient are only lazy classes ,carrying the data and don't have any behavior.
- Separation of concerns the program is not divided into small, logical, self explanatory classes. The classes mainly do too much .Example of the class doing too much is DataPointGenerator, which basically calculates all data needed writes them to the Arff file but also saves the file to the disk.
- Avoid god classes The DataPointGenerator is definitely a god class here. It has about 50% of functionality of whole application embedded in it.
- Long method smell The DataPointGenerator has the methods that involve many conditional statements, special in the part when calculating attributes values, this suggests that the method should be split up in the smaller tasks (methods)..
- Distribute system intelligence The breach of this principle is closely related to the Avoid god classes and Large class smell. I clearly didn't divide my system intelligence well enough. All system intelligence is concentrated in one class (DataPointGenerator).
- Coupling and cohesion There is quite a strong coupling between classes (eg. DataPointGenerator and ModelBuilder where ModelBuilder contains a pointer directly to a concrete class DataPointGenerator which provides the required behaviour).
Improved design
Following the smells and basic design principles I have come up with my improved design. The Uml follows:
New design analyze
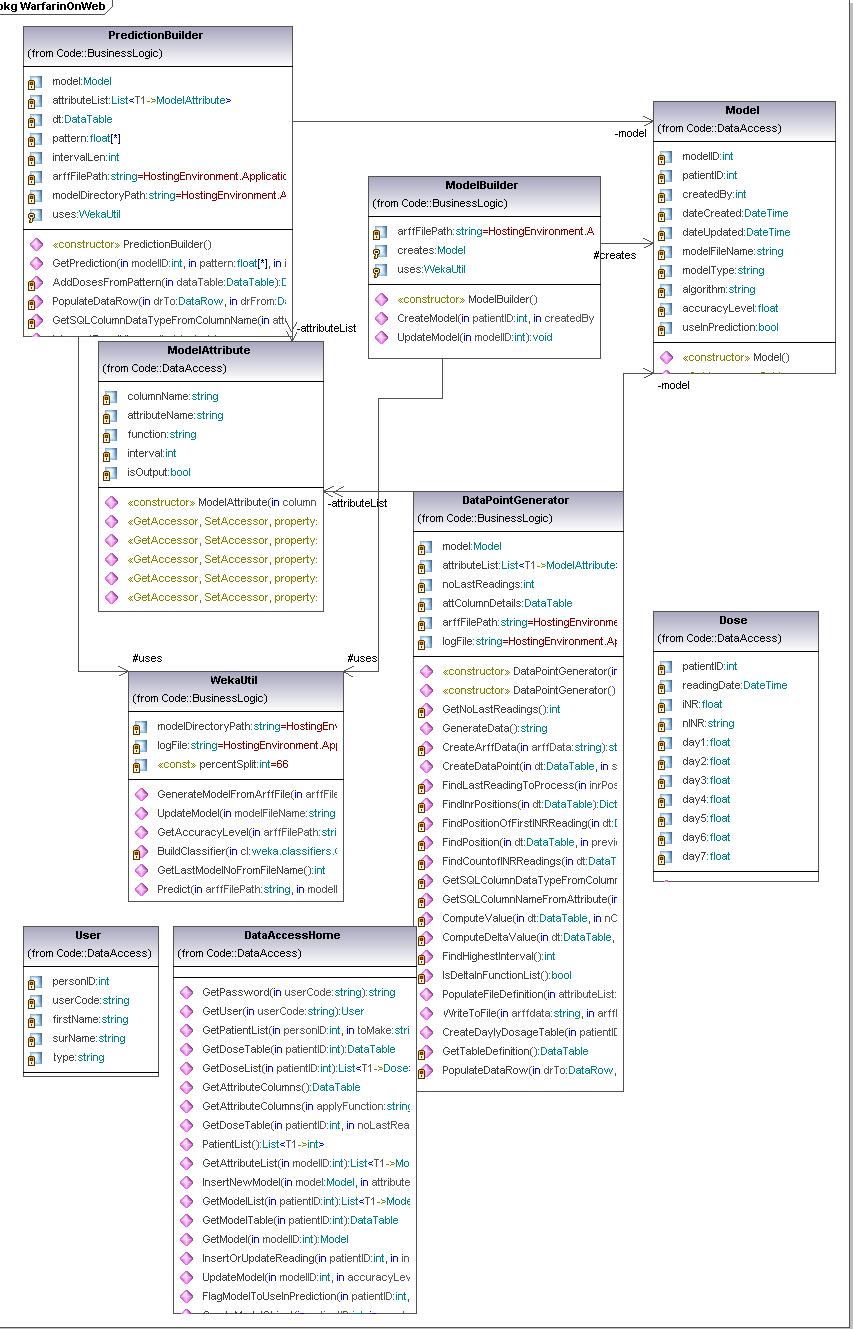
Getting to the stage when I am comfortable with my new design involved many hours of grappling with some important design issues and decisions. In that process I have been through close to 10 different UML designs. Although I tried to follow the design best practices and maxims, sometimes compromises had to be made in terms of the design approach in the given situation. Not an easy task. I will first name all my classes and briefly explain what their roles are. Then I will talk about principles that I strongly followed, conflicts that I encountered, and how and why I made the decisions that I made. Before I list all my classes I want to point out that my UML has most important fields, but because I tried to keep UML complete but not cluttered I left out constructors and public properties (getters and setters). I also removed all fields that are already implied by relationships.
Classes used in the new design
- Patient - this class is used to present the patient with it's history. It is mostly data class, used to carry data relevant to a patient. Saving the patient, updating the patient or finding the patient (or list of patients) are all implemented in DataAcessor class, due to layered architecture that I wanted to achieve here. Will talk about this more in the next section.
- BloodTest - this class is used to carry data related to regular blood tests that person takes. Every person could have many of those.
- Dose - this abstract class presents one dose on the specific date. People take Warfarin once a day , the time was not relevant as per initial requirements but this could change so the data type used here is DateTime (not just date). Each dose has two concrete subclasses ActualDose and PrescribedDose. Ideally those should be the same but there is a good chance to be different due to human error.
- Doctor - this class presents a doctor that is our design same as a user that logs to the website. This is the reason for having a user code field. The doctor creates a PatientModel and Prediction.
- PatientModel - this class represents a model that can be used to predict INR reading. I is responsible for constructing a model, saving it and updating it.
- Prediction - this class is responsible of making a prediction based on the model selected by a user. It interacts with Arff file and model and based on the dose pattern that doctor find appropriate makes a prediction. It also makes a calls to WekaUtil in order to utilize matching learning algorithms.
- Algorithm - this class is again data class used to present one ML algorithm. Every model is build by using one.
- ModelCreator - this class has responsablity of iterating through selected list of algorithms for a selected list of attributes and create as many models as there are algorithms.
- ArffFile - this class represents the Arff file containing patients history data transformed into data points as per selected model attributes. It is responsible for creating the file with all it's data and saving it to the disk.
- HeaderWriter - this class is used to create arff file header definition.
- AttributeDataWriter - this is an abstarct class subclassed into AttributePredictionDataWriter and AttributeModelDataWriter. Both make data points in arff file where the first one does it for making a prediction while the subsequent one makes it for model creation.
- ModelAttribute - this is an abstract class responsible of representing a model attribute created by a user.Important fields here are attributename but also the columnname which reporesents the name of the data column(database column) whose data this attribute should be based on. This column should match one of the patient's class fields .Value represents the calculated model attribute value for a given patient and given bloodtest date.
- ClassAtribute - this is ModelAttribute's subclass. It can be nominal or numeric but also differs if is made for prediction or model building purposes.
- CalculatedAttribute - this a MOdelAttribute sublass that usesfunction passed to it to invoke a sutable strategy to claculate the attribute value.
- StaticAttribute - this is the concrete class, ModelAttribute subclass used to give back the information on the patient that doesn't need calculation (eg.dob,gender...)
- ICalculationStrategy - this is an interface used to specify the strategy signiture.. Is then implemented by two concrete strategies.
- AggregateStartegy - this class reporesents a concrete strategy taht implements ICalculationStrategy used to calculate modelAttribute value when there is an aggregate function to be used (sum,min, max,avg).
- DeltaStrategy - a concrete class that implements ICalculationAttribute and calculates delta value between sum of column data for two subsequent blood test readings.
- AttributeColumnDefinition - this class hold all necessary information on patient's database column. It is a datacolumn that is open for user to select when designing a model attribute. Therefore we need to know it's database characteristics but also which data field in Arff file this data field match to.It is used by HeaderWriter when creating the arff file header but also by AttributeDataWriter
- WekaUtil - this class is used to make all API calls to WEKA algorithms.
- DataAccessor - this class calls SQL stored procedures for retrieving, updating and persisting the data classes.
Design decisions
- ActualDose and PrescribedDose classes are currently empty. There is nothing to distinguish them at this point so it might look odd to have them .Few principles work against them You ain't gonna need it , Eliminate irrelevant classes and Lazy class smell. I decided to include them because Model the real world principle would recommend to have them as the difference between the two are bound to happen in real life but also because I will need to do the investigation on how much actual dose can vary to prescribed dose without causing an undesirable INR reading.
- PatientModel object can be made based on a single patient history and in that case would only make sense to be used by that patient, but if it was made on multi patient history can be used in prediction for any patient. So the principle Model the real world would suggest here for me to have two subclasses here SinglePatientModel and MultiPatientModel and make my model more extensible , but I think that You ain't gonna need it probably overrules this in this case. I think that having a list of patient field on the PatientModel that can have either one element in a case of single model or many patients in a case of a multi model would be sufficient for now. If I ever need to find if a patient's history was included in the model I can add the method IsIncluded() that would do that kind of testing for me.
- I had a problem with finding the right names for a few classes. One is PatientModel class. I initially named it Model but decided that the name is too generic. I was thinking about Classifier as another option, but only the person that is familiar with the domain can understand it otherwise the term could be too vague. So I made a decision to call it PatientModel since it is made on the bases of patient data and it is made for the purpose of predicting the patient’s blood test, so it is all to do with the patient. The other one is the ModelAttribute class. It is also pretty vague and not descriptive but is used in domain and I think I have used the term quite a lot in my discussion of how the system works that even a new comer can find it comprehensive.
- Initially in my old design I had a class named ModelBuilder that iterated through the chosen algorithms and made many new models based on one arff file ( so based on the same patient history) .I initially thought that PatientModel itself might do this by changing the algorithm field when iterating through the list of algorithms selected and since all the other characteristics of the PatientModel would stay the same I would just call WekaUtil from PatientModel to create as many model files as there are algorithms. But PatientModel shouldn't really take care of this. This would break Single Responsibility Principle and One key abstraction. So I introduced ModelCreator and pulled only CreateModel method out into this new class. This method should create a completely new model each time (call the PatientModel constructor) and set the correct parameters. The PatientModel would then have a responsibility of representing a model and not creating lots of new ones.
- I have some data classes in my design (e.g. patient and algorithm) so I think I should justify them. The Lazy class smell and Data class smell, as well as the Anaemic domain model anti-pattern apply here. However, I think that Model the real world was important in this instance and it is highly likely that more behaviour will be added to the classes later.
- I already mentioned when I was criticising my initial design that it didn't follow Open-closed principle because it didn't allow introducing a new way of calculating the modelAttribute without making some serious changes to DataPointGenerator. In order to change this and make this design extensible I introduced Strategy pattern when it comes to calculating the attribute values. Since I have 3 different types of ModelAttributes and only CalculatedAttribute needs a computationStrategy, I made CalculatedAttribute a context for my strategies. Depending on the selected calculated attribute (the function used) an appropriate strategy would be instantiated. I also made strategies Singletons to make sure that I am not needlessly instantiating a new strategy objects for each datapoint and for the cases when few attributes are using same strategy (and the only difference for example is interval to apply aggregate function to).
- There is possibility for implementing one more design pattern in this design. The Template pattern. I thought that I could use it in the area of AttributeWriter where I need to make a different files depending on weather the file is made for prediction or for building a model. I could have an abstract class that would define the steps in writing header and data to the arfffile and have methods like
WriteHeading()
WriteAttributeValues()
and then implement those in subclasses differently. I decided not to do this since there are not as many different steps in writing data to the file (it is only one, header would be the same in both cases, and it is a common knowledge that if pattern doesn't help with code flexibility a great deal it is better not to implement it since it is adding a complexity.
- When it comes to encapsulation I investigated couple of relevant but conflicting design principles: Hide data within its class and Avoid protected data.I also read Encapsulation boundary research and decided to use object encapsulation. I am aware that this is going to break Riel's heuristic of hiding data within its class, but it only seemed natural to go with object encapsulation and allow subclasses to access parent's fields by making them protected. Those fields would remain private for the rest of the world. Since my project is done in C# protected fields are available to use.
- I would like to explain my decision on putting all the database methods into one common class called DataAcessor. I am aware that this is breaking a very important OO principle Keep related data and behavior in one place, but this would on the other hand go well with other conflicting principle Separation of concerns since writing to the database is not really the functionality of the Patient object or PatientModel object for example, so this is something that needs to be separated out. I decided to obey Separation of concerns here for one more reason. Having a database access all in one place makes the data source change doable by changing only one class. This would mean that I implemented n - tier architecture (having presentation layer, domain logic layer , data access layer and physical database layer). If my application would to grow much bigger then what it currently is, my class DataAccessor would become too big and I would end up with Large class smell but for now it seems like a good decision backed up with the reasoning that we don't expect this to grow very much at all.
Design extensibility
Easy extensibility and flexibility of the design were high on my priority list when I started improving my initial design. I found really crucial to be able add new types of model algorithms to the process of "learning" a model. Those could come form collecting a new data on the patients that we thought could make a difference to the prediction results ( eg. we could decide to record and process blood pressure) but also we could think of better ways of transforming patient's history (eg. like using the data ratios). I think that the design that I had initially would require for every class to be changed to achieve that request. This is naturally not desirable thing to have. In the new design I could achieve adding a new patient data by adding a new record to the XML file that is used to feed AttributeColumnDefinition class. This would bring this new field to the user selection screen. I would also need to add that same field to the patient class. To add a new way of computing any numeric values stored on a patient , I would only need to add a new strategy with CalculateValue method implementation. By doing this I am not changing the existing classes, I am only extending the design by adding a new class that is inheriting from abstract class that didn't need to change. This makes Open-close principle and Stable abstractions principle fully implemented.
Summary
To summerize I am think taht this project was very useful. The work that I have done on redisagning my code made me rethink and question every single idea that I initially had, which overallhelped me grow in OO sense. I have started with a very messy code that has most of the required functionality but would be an nightmare to maintain and extend. I think that the quote that we shouldn't be programming just to achieve the functionality but for other programmers to continue the work on and extend, makes a lot of sense . If everyone would think in that way we would not just spend less time doing maintaince but would save the money spent on the projects overall. I expect that anyone working on my project from this point on should find it easy to work with and quite intuitive .
Code
This is my inital code Media:WarfarinPredictor.zip
Note: all the weka.dlls that are used to make API calls to weka's machine learning algorithms are taken out due to the size ( over 8MB)
Code for the improved design is coming soon.